Group Testing Method Developed for COVID-19
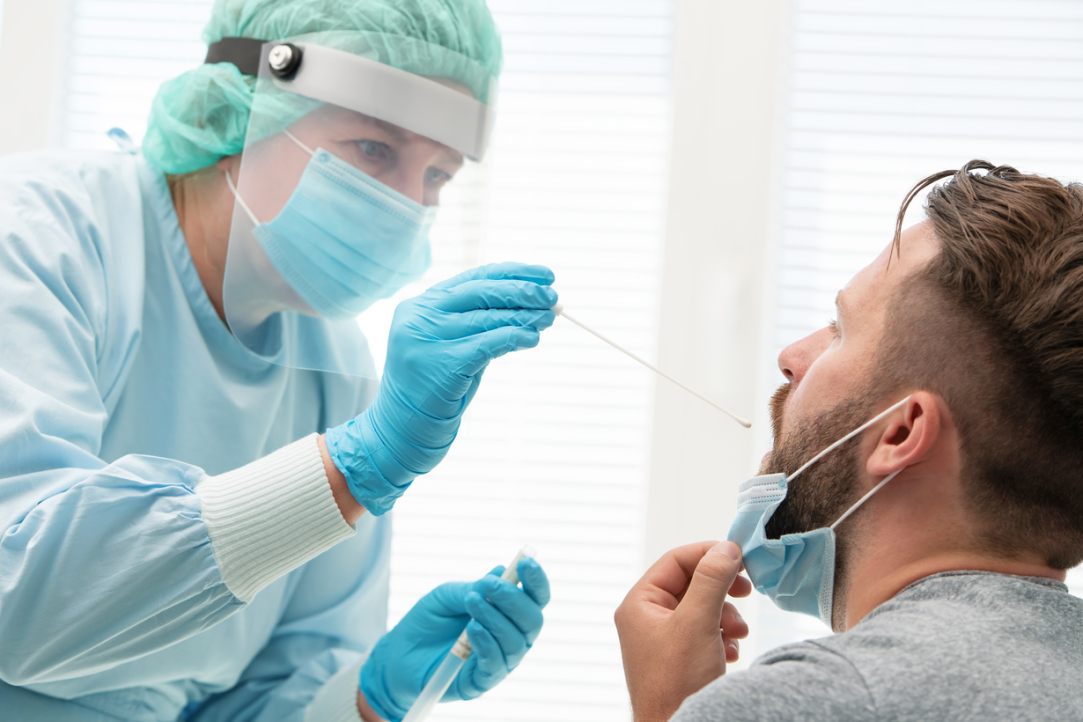
Researchers Mario Guarracino from the HSE Laboratory of Algorithms and Technologies for Networks Analysis in Nizhny Novgorod and Julius Žilinskas and Algirdas Lančinskas from Vilnius University, have proposed a new method of testing for COVID-19. This group method allows results to be obtained 13 times faster as compared to individual testing of each sample. The research paper was published in the journal Scientific Reports.
The COVID-19 pandemic has already affected millions of people from over 200 countries. The rapid virus expansion demonstrated how fast such infections can spread in today’s globalized world. At the beginning of the pandemic, when little was known about the virus and vaccines had not yet been developed, it was possible to slow its spread only by means of limiting the population’s mobility. Almost everyone around the world went through various lockdowns and periods of isolation. If large groups of people can be tested quickly, the restrictions can be less strict and more effective at the same time, believe the authors of the paper 'Pooled testing with replication as a mass testing strategy for the COVID-19 pandemics.’

Present COVID-19 testing solutions are based on the extraction of RNA from patients using oropharyngeal and nasopharyngeal swabs, and then testing with real‑time PCR for the presence of specific RNA filaments identifying the virus. The speed of this approach is limited by the availability of reactants, trained technicians and laboratories.
One way to speed up the testing procedures is group testing, where the swabs of multiple patients are grouped together and tested. The swabs from groups that return a positive result are then tested individually in order to detect specific COVID-19 positive patients. This approach helps decrease the number of tests twofold or more (depending on the spread of the disease) as compared to individual testing of each swab.
For example, suppose 96 samples should be tested and pools of up to 12 samples are possible. In individual testing, 96 tests are necessary. In pool testing, 8 pools of 12 samples are taken and testing is performed. If the result of one pool is positive, then additional 12 individual tests are needed. If two or three groups return a positive result, 24 or 36 additional tests are required, which, together with the first eight tests, will mean a decrease in the number of tests from two to five times as compared to individual testing.
The researchers believe that the number of tests can be decreased by optimizing the size of groups that takes into account the total number of swabs and the forecasted number of infected individuals. As the number of infected individuals increases, the possibility of saving swabs decreases but is still about 40% in the event of an incidence of 100 positive samples per 1,000, and 18% for an incidence of 200 per 1,000.
There are ways to optimize group testing, such as choosing the optimal group size based on the total number of swabs and the projected level of disease spreading. Another is the binary splitting method, in which a positive group is split into halves and is tested again, until individual positive swabs are detected. The second method, however, is very time-consuming, which decreases its attractiveness during a pandemic.
In addition, to optimize group testing, transposition-based replication is used: after grouping the swabs, researchers form additional control groups from the same swabs and test them together with the main groups. This helps further cut the number of tests, and if the disease levels are low, it also helps to detect positive swabs in one step, which speeds up the testing considerably.
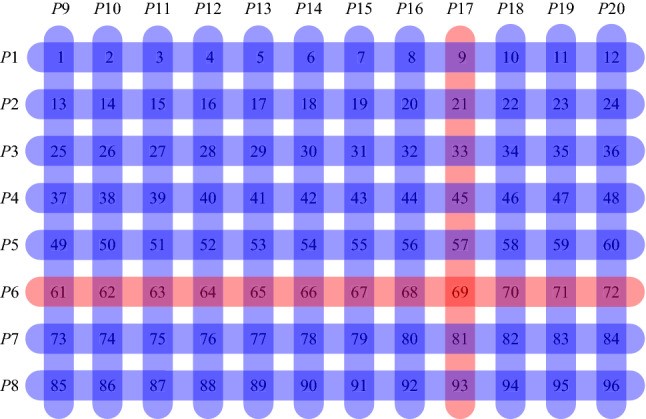
However, this method does not allow for experimenting with group sizes to detect the optimal group size under specific conditions. Researchers from HSE University and Vilnius University suggested OptReplica technology, which uses a more complicated algorithm of swab grouping in key and control groups and helps decrease the number of control groups. In addition, the algorithm helps calculate the optimal group size for the present number of swabs and the forecasted level of disease spreading.
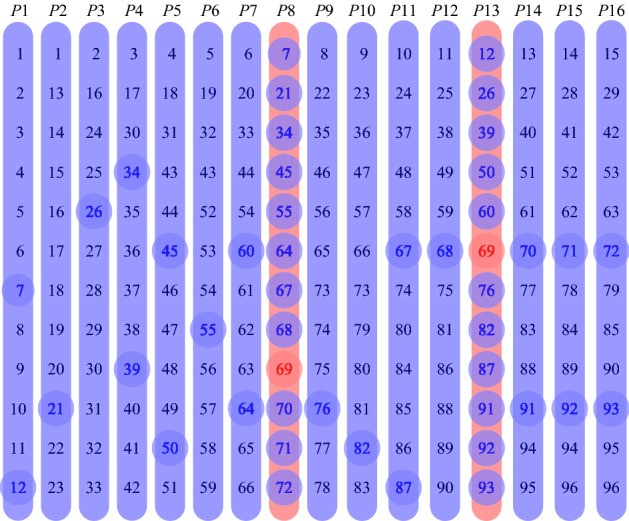
The authors conducted experimental research on samples of 96 and 384 swabs, carrying out 100 randomized tests for each sample size, and compared the effectiveness of transposition-based replication and OptReplica method for different levels of disease incidence. The studies have shown that if the optimal size of groups is chosen, OptReplica is more effective than transposition-based replication. In cases with low incidence, the use of OptReplica, a 13x average reduction of tests can be achieved compared to individual testing without time delay.
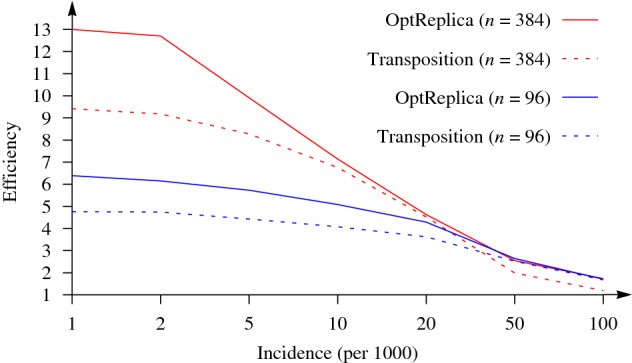
‘Our simulations are actually proving that using this optimization replication strategy is always advantageous and, even in case of high spread of the disease (10% or 20% of positives in the population), we are still competitive with individual testing strategy’, explained Mario Guarracino, Chief Research Fellow of the Laboratory of Algorithms and Technologies for Networks Analysis.
The authors of the new technology suggest using it for asymptomatic populations with seemingly low incidence of coronavirus cases, where it will help detect the infected individuals at a maximum speed with a minimum number of tests, and timely apply the quarantine measures in order to prevent spreading of the disease. In regions with disease incidents over 50 cases per 1,000 tests, the authors suggest using other methods of group replication, or testing without replication.

